MBH-Seg25: Multi-class Brain Hemorrhage Segmentation in Non-contrast CT
Leaderboard
Check the leaderboard here:Leaderboard
Challenge Day
Date & Time: Korea Standard Time (UTC+9) - September 27th, 2025, 8:00 AM - 10:00 AM
Location: On-site (IBS-3F-R1-R3)
Zoom Recording:https://uni-sydney.zoom.us/rec/share/TaKP5hGzlCy74LQhUVzs3lXuht7n1U19n4i64lBOI_yBx6m4lxRcA9Cszx-O3gl8.9eGOgwXWeakvxzGK (Passcode: $Cc@N2n%)
Zoom Recording:https://uni-sydney.zoom.us/rec/share/TaKP5hGzlCy74LQhUVzs3lXuht7n1U19n4i64lBOI_yBx6m4lxRcA9Cszx-O3gl8.9eGOgwXWeakvxzGK (Passcode: $Cc@N2n%)
Schedule
| Time | Presenter/Event |
|---|---|
| 8:00-8:15 | Welcome & Overview |
| 8:15-8:25 | Jesús David González, Vall d'Hebron Institut de Recerca |
| 8:25-8:35 | MRNN, Eindhoven University of Technology |
| 8:35-8:45 | Lizhi He |
| 8:45-9:00 | AMC-Lab, Asan Medical Center |
| 9:00-9:15 | CLAIM, Charité – Universitätsmedizin Berlin |
| 9:15-9:30 | Dolphins, University College London |
| 9:30-9:45 | Rana Shahzaib, University of Alberta |
| 9:45-10:00 | Closing Ceremony |
Overview
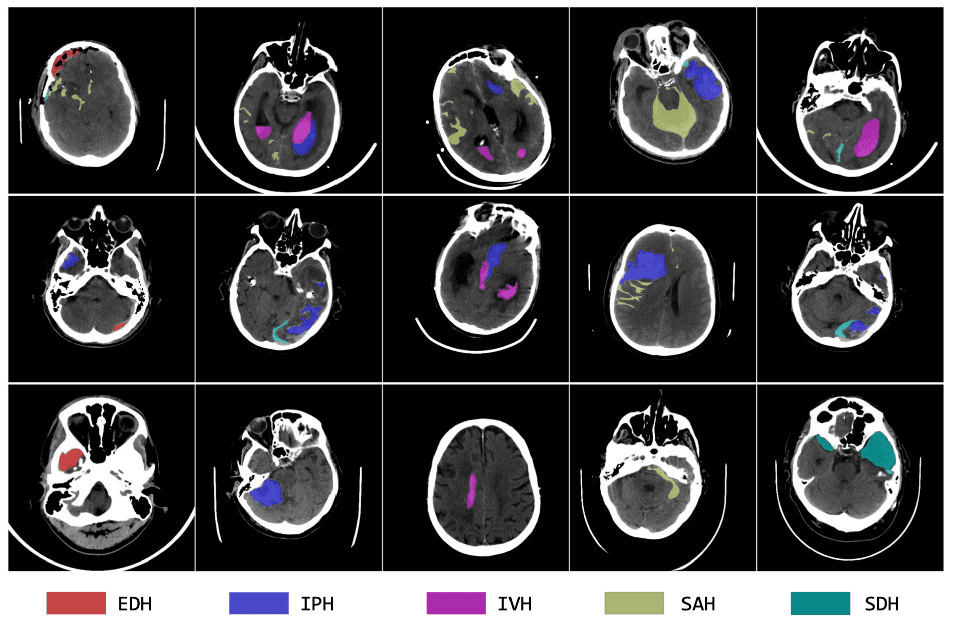
We warmly invite you to participate in the second edition of the MICCAI MBH-Seg25 Challenge on multi-class brain hemorrhage segmentation from non-contrast CT scans. Accurate segmentation of different hemorrhage subtypes—such as subdural, epidural, intraparenchymal, and subarachnoid—is critical for timely diagnosis, treatment planning, and clinical decision-making. Each type of hemorrhage carries distinct prognostic and therapeutic implications, underscoring the need for precise and reliable delineation. Currently, diagnosis primarily depends on expert radiologists interpreting non-contrast CT scans—a process that is time-consuming and prone to inter-observer variability. This challenge aims to transform traditional workflows by promoting the development of advanced AI-driven segmentation methods that enable faster, more consistent, and more accurate diagnosis, ultimately reducing mortality and improving patient outcomes.Building on the success of last year’s challenge, MBH-Seg25 introduces new settings that better reflect real-world clinical scenarios and data constraints:
1. We incorporate multi-rater annotations to capture variability among experts, encouraging the development of algorithms that can handle disagreements and uncertainty in medical data.
2. We provide extra weakly-labeled cases with case-level labels to simulate scenarios where detailed annotations are unavailable, promoting research into methods that make the most of limited or less granular data.
Schedule
| Event | Timeline (Anywhere on Earth) | ||||
|---|---|---|---|---|---|
| Training Data Release | June 15, 2025 | ||||
| Validation Data Release | August 10, 2025 | ||||
| Testing Phase Submission (with Method Description) | August 25, 2025 – September 2, 2025 | ||||
| Notification of Invited Presentations | September 7, 2025 | ||||
| Final leaderboard announcement | September 14, 2025 | ||||
| Challenge Day and Awards Ceremony | September 27, 2025 | ||||
Dataset
Evaluation
For evaluation, we follow [4] to do the evaluation for multi-rater medical image segmentation. In general, we evaluate the segmentation results from two aspects:
- Diverse performance: we use the GED score, Dicesoft score, and two set-level metrics DiceMatch and DiceMax to compare the similarity between the results set and label set.
- Personalized performance: we compute the Dice score for each individual expert and then use their mean value as the final metric.
We will use a combination score of these metrics for the final ranking. You can find the test script at here
During the testing phase, each team is allowed a maximum of three submissions. The final test set will not be publicly released. Instead, participants are required to submit the following:
- Model weights
- Testing scripts
- A README.md file with clear instructions on how to execute the testing pipeline
- A comprehensive Method Description document, using the MICCAI 2025 paper template. Manuscripts can be up to 8 pages (text, figures and tables) plus up to 2 pages of references.
The final leaderboard rankings will be determined based on the results from the testing-stage evaluation. Additionally, the submitted Method Description will be taken into consideration when selecting invited presentations and awards.
Submission
ClosedPrizes
1. Cash Awards:First Prize: $800
Second Prize: $400
Third Prize: $200
2. Certificates for the top-rank teams, and the teams with the careful method description.
3. The top teams will be invited to give oral presentations at the MICCAI 2025 Event.
Organization
Organizers
•Dongang Wang, Postdoctoral Researcher, Sydney Neuroimaging Analysis Centre, University of Sydney, Australia•Yutong Xie, Assistant Professor, Mohamed bin Zayed University of Artificial Intelligence, The United Arab Emirates
•Chenyu Wang, Associate Professor, Sydney Neuroimaging Analysis Centre, University of Sydney, Australia
•Zihao Tang, Postdoctoral Researcher, University of Sydney, Australia
Clinical Support
•Minh-Son To, Medical Practitioner, Flinders Health and Medical Research Institute, Flinders University, Australia•Laurent Letourneau-Guillon, Neuroradiologist & Associate Professor, University of Montreal & Centre de Recherche du CHUM (CRCHUM), Canada
•Advait Pandya, Senior Resident Medical Officer, Royal Prince Alfred Hospital, Australia
•Jashan Manjit Dugal, MD Research Student, University of Notre Dame, Australia
Contributors
•Siqi Chen, Research Assistant, Mohamed bin Zayed University of Artificial Intelligence, The United Arab Emirates•Jiayu Niu, Undergraduate Student, South China University of Technology, China
•Xuechenxiao Cao, Undergraduate Student, South China University of Technology, China
•Mohammad Yaqub, Associate Professor, Mohamed bin Zayed University of Artificial Intelligence, The United Arab Emirates
•Rao Muhammad Anwer, Assistant Professor, Mohamed bin Zayed University of Artificial Intelligence, The United Arab Emirates
Advisor
•Michael Barnett, Professor, University of Sydney. Australia•Qi Wu, Assoiate Professor, Australian Institute for Machine Learning (AIML), University of Adelaide, Australia
•Jian Chen, Professor, South China University of Technology, China
Organized By








Terms and Conditions
1. We only consider automatic segmentation methods for the final ranking.
2. No additional data is allowed. We will require all participants to provide extensive documentation on their development processes, including data and methods used. Top-ranked participants must submit their training codes and checkpoints for verification. Publicly available pre-trained models are allowed, ensuring a level playing field and transparency in the competition's evaluation process.
3. Independent Publications: Participating teams are allowed to publish their results separately. However, this should be done in a manner that respects the collective efforts of the challenge.
2. No additional data is allowed. We will require all participants to provide extensive documentation on their development processes, including data and methods used. Top-ranked participants must submit their training codes and checkpoints for verification. Publicly available pre-trained models are allowed, ensuring a level playing field and transparency in the competition's evaluation process.
3. Independent Publications: Participating teams are allowed to publish their results separately. However, this should be done in a manner that respects the collective efforts of the challenge.
Acknowledgments and Citations
We express our gratitude for the significant contribution of the RSNA Intracranial Hemorrhage Detection Challenge Dataset.
1. Wu, Biao, Yutong Xie, Zeyu Zhang, Jinchao Ge, Kaspar Yaxley, Suzan Bahadir, Qi Wu, Yifan Liu, and Minh-Son To. "BHSD: A 3D Multi-class Brain Hemorrhage Segmentation Dataset." In International Workshop on Machine Learning in Medical Imaging, pp. 147-156. Cham: Springer Nature Switzerland, 2023.
2. AE Flanders, LM Prevedello, G Shih, et al. Construction of a Machine Learning Dataset through Collaboration: The RSNA 2019 Brain CT Hemorrhage Challenge. Radiology: Artificial Intelligence 2020;2:3.
3. Anouk Stein, MD, Carol Wu, Chris Carr, George Shih, Jayashree Kalpathy-Cramer, Julia Elliott, kalpathy, Luciano Prevedello, Marc Kohli, MD, Matt Lungren, Phil Culliton, Robyn Ball, Safwan Halabi MD. (2019). RSNA Intracranial Hemorrhage Detection. Kaggle. https://kaggle.com/competitions/rsna-intracranial-hemorrhage-detection
4. Wu, Y., Luo, X., Xu, Z., Guo, X., Ju, L., Ge, Z., Liao, W., Cai, J.: Diversified and personalized multi-rater medical image segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). pp. 11470–11479 (June 2024)
2. AE Flanders, LM Prevedello, G Shih, et al. Construction of a Machine Learning Dataset through Collaboration: The RSNA 2019 Brain CT Hemorrhage Challenge. Radiology: Artificial Intelligence 2020;2:3.
3. Anouk Stein, MD, Carol Wu, Chris Carr, George Shih, Jayashree Kalpathy-Cramer, Julia Elliott, kalpathy, Luciano Prevedello, Marc Kohli, MD, Matt Lungren, Phil Culliton, Robyn Ball, Safwan Halabi MD. (2019). RSNA Intracranial Hemorrhage Detection. Kaggle. https://kaggle.com/competitions/rsna-intracranial-hemorrhage-detection
4. Wu, Y., Luo, X., Xu, Z., Guo, X., Ju, L., Ge, Z., Liao, W., Cai, J.: Diversified and personalized multi-rater medical image segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). pp. 11470–11479 (June 2024)
Contact Us
Please contact us if you have any enquires through our Google Form